Summary
The goal of precision medicine is to tailor medical treatments for patients based on their personal characteristics and responses to a specific treatment. Genome-wide association studies and pharmacogenomics are tools that may help clinicians determine how best to treat patients, such as those with breast cancer, based on genetic factors and phenotype.
- precision medicine
- genome-wide association study
- pharmacogenomics
- breast cancer
- aromatase inhibitors
- genomics
- endocrinology, diabetes & metabolism
James N. Ingle, MD, Mayo Clinic, Rochester, Minnesota, USA, presented the first of 2 Presidential Plenary sessions focused on the value of precision medicine in the endocrine treatment of breast cancer (BC).
Dr Ingle began by reviewing the National Research Council’s definition of precision medicine as “the tailoring of medical treatment to the individual characteristics of each patient” and “the ability to classify individuals into subpopulations that differ in their susceptibility to a particular disease or . . . in their response to a specific treatment” [National Resource Council. Toward Precision Medicine. USA: National Academies Press; 2011: 125]. He then went on to discuss how pharmacogenomics—the study of genetic variation across the entire genome in drug response—influences precision medicine.
The clinical goals of pharmacogenomics are 3-fold: (1) to select patients who are most likely to respond to a drug, (2) to maximize the efficacy of a drug, and (3) to mitigate adverse drug reactions. As well, pharmacogenomics provides researchers with a methodology to understand the mechanisms of a given drug on a specific population.
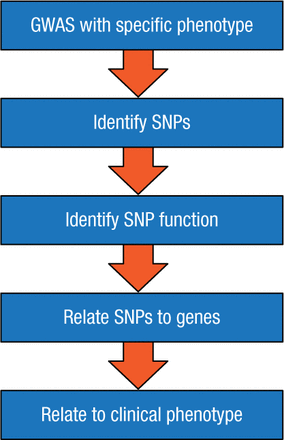
Dr Ingle outlined an approach to achieve these goals, utilizing a genome-wide association study (GWAS; Figure 1), which scans the complete genomes of many people to find variability related to a specific phenotype or disease. A GWAS typically focuses on the association between single-nucleotide polymorphisms (SNPs)—changes in a single sequence of DNA—and traits that are characteristic of major diseases. For example, GWAS has enabled scientists to map genome-wide differences in estrogen receptor (ER)-α binding and to predict which women with ER-positive BC are likely to have distinct clinical outcomes [Ross-Innes CS et al. Nature. 2012].
Flowchart of a GWAS Study
GWAS, genome-wide association study; SNP, single-nucleotide polymorphism.
Reproduced with permission from JN Ingle, MD.
Dr Ingle described how GWAS studies have informed clinical research regarding the medical treatment of postmenopausal women with ER-positive BC [Liu M et al. Mol Endocrinol. 2013]. Using GWAS, researchers were able to observe a statistical association between SNPs in the TSYPL5 gene with fluctuating concentrations of estradiol and aromatase, an enzyme responsible for a key step in the synthesis of estrogen. According to Dr Ingle, these results represent a new mechanism for the control of aromatase and estrogens in postmenopausal women and may offer insights as to why some women can tolerate aromatase inhibitors (AIs) and others cannot.
Dr Ingle then went on to discuss the fact that many women experience arthralgias and myalgias when undergoing BC treatment with AIs and that this is a common reason why women stop their treatment. An earlier GWAS had identified an SNP near the 3′ end of the T-cell leukemia 1A (TCL1A) gene that was associated with musculoskeletal pain in women who were taking AIs to treat their BC [Ingle JN et al. J Clin Oncol. 2010]. He then reviewed results from a GWAS that genotyped DNA from the cell lines of a total of 300 healthy European-American, African-American, and Han Chinese-American women [Liu M et al. Breast Cancer Res. 2012]. The results suggested that increased expression of the TCL1A gene upregulated expression of interleukin-17 receptor A, which is an indicator of inflammation often seen in patients with rheumatoid arthritis (RA). Results from another GWAS used 300 different lymphoblastoid cell lines cultured in increasing concentrations of estradiol [Ho M et al. Clin Pharm Ther. 2014]. These results showed that TCL1A-mediated regulation of chemokine receptor 6 (CCR6)—a cytokine associated with the development of RA—was SNP dependent. According to Dr Ingle, this raises the possibility that postmenopausal women known to have a variant TCL1A SNP might be more likely to develop arthralgias similar to those experienced by people with RA. If this could be pharmacologically manipulated perhaps the arthralgias could be minimized, which might allow patients to continue their treatment with AIs.
Dr Ingle closed his talk by highlighting genetic research regarding the use of selective estrogen receptor modulators (SERMs) to prevent BC. To date, the two largest SERM BC prevention trials are the National Surgical Adjuvant Breast and Bowel Project (NSABP) P-1 trial of tamoxifen [Fisher B et al. J Natl Cancer Inst. 1998] and the NSABP P-2 trial that compared raloxifene with tamoxifen [Vogel VG et al. Cancer Prev Res (Phila). 2010]. Combined, these studies involved > 32 000 women and were the basis of the 2 drugs being approved by the US Food and Drug Administration to prevent BC [Ingle JN et al. Cancer Discov. 2013]. While these trials confirmed that 5 years of treatment with raloxifene and tamoxifen could reduce the recurrence of BC by one-half, the drugs are not routinely used for this indication. This is due in part to the high number of patients needed to treat to prevent one case of BC and because the SERMs can be associated with worrisome side effects.
Dr Ingle and his colleagues performed a GWAS that included 592 cases and 1171 controls from the NBASP P-1 and P-2 trials [Ingle JN et al. Cancer Discov. 2013]. A ZNF423 SNP variant on chromosome 16 was associated with lower breast cancer risk (OR = 0.7), and these variant SNPs were found to be an estrogen-inducible BRCA1 transcription factor. A CTSO SNP variant on chromosome 4 was associated with an increased risk of developing BC (OR = 1.42) and was found to disrupt the estrogen receptor element. The combined odds ratios for these 2 sets of SNPs suggest a broad range of relative odds ratios for the development of BC for women on SERM therapy for 5 years. Dr Ingle highlighted the fact that both the CTSO and the ZNF43 SNPs appear to be estrogen inducible and regulate estradiol-dependent induction of BRCA1. He also emphasized that SERMS can reverse that SNP-dependent expression.
In summary, Dr Ingle emphasized that a GWAS is the starting point for a process that examines how an SNP works, how it relates to specific genes, and how these genes then influence the effect of the drug on a clinical phenotype. While pharmacogenomics studies have identified new biology and have substantial potential to provide clinical benefit, further work is needed to validate the clinical relevance and value of this approach.
- © 2015 SAGE Publications