Summary
This article discussed the curability of luminal A and luminal B breast cancer subtypes, pharmacogenetics/pharmacogenomics to optimize endocrine therapy, new targeted agents in luminal breast cancer, as well as a review of whole genome sequencing for luminal-type breast cancer [Ellis MJ et al. Nature 2012].
- Oncology Genomics
- Breast Cancer
- Adjuvant/Neoadjuvant Therapy
Angelo Di Leo, MD, PhD, Hospital of Prato, Instituto Toscano Tumori, Prato, Italy, discussed the curability of luminal A (LUM A) and luminal B (LUM B) breast cancer subtypes. Hormone receptor (HR)-positive breast cancers are separated via molecular subtyping into LUM A (higher prevalence and less aggressive) and LUM B (higher grade, increased proliferation rates, and poorer prognosis). Immunohistochemical assessment of the proliferative marker Ki67 is another possible mechanism to differentiate between the subtypes; however, the false-positive and false-negative rates are high. Development of mechanisms to differentiate between subtypes is critical so that appropriate prognosis information and treatment options can be identified.
Endocrine therapy alone is generally recommended for LUM A, although tumor dormancy and late recurrences (beyond 10 years) are potentially problematic. The optimal choice and duration of endocrine therapy remain largely unknown. LUM B is associated with more aggressive disease; therefore, it is generally treated with both endocrine therapy and chemotherapy. However, this approach is not always effective. Additional treatment options may include identification and targeting of other important pathways active in LUM B.
The Breast Cancer Trials of Oral Everolimus-2 [BOLERO-2; NCT00863655] study examined the effect of targeted agents in addition to endocrine therapy. Combination therapy (exemestane plus everolimus) improved survival compared with exemestane alone but was associated with more adverse events and a higher dropout rate [Baselga J et al. N Engl J Med 2012]. A Phase 2 randomized trial of tamoxifen alone versus tamoxifen plus everolimus in patients with advanced breast cancer who were previously exposed to nonsteroidal aromatase inhibitors (AIs; n=111) found that the most benefit was experienced by patients who had an initial response to AI therapy [Bachelot T et al. J Clin Oncol 2012]. Currently, there is no biologically driven strategy for the use of tamoxifen versus AI in the adjuvant setting. The only factor supporting treatment decisions is risk of relapse.
James N. Ingle, MD, Mayo Clinic, Rochester, Minnesota, USA, discussed pharmacogenetics/pharmacogenomics (PGx) to optimize endocrine therapy. PGx is the study of the role of inheritance in individual variation in drug response phenotypes. Clinically, endocrine therapy produces variability between patients in terms of clinical response, adverse events, and end-organ effects. For example, there is a striking difference in musculoskeletal events associated with AI therapy, as well as in the incidence of deep vein thrombosis, hot flashes, and lipid effects. Most single-gene studies to date have focused on the P450 enzyme CYP2D6 which mediates the conversion of tamoxifen into endoxifen; however, results have been inconsistent, likely due to flawed retrospective studies. Additional studies have examined polymorphisms in Phase 2 enzymes involved in tamoxifen/endoxifen metabolism (UDP glucuronosyltransferase and sulfotransferases), as well as estrogen receptors 1 and 2; however, results are preliminary.
Prof. Ingle and colleagues conducted the largest pharmacogenomics genome-wide association study (GWAS) in high-risk women receiving tamoxifen or raloxifene (National Surgical Adjuvant Breast and Bowel Project [NSABP] P-1 and P-2 Prevention Trials) and identified single nucleotide polymorphisms (SNPs) associated with development of breast cancer and SNP-dependent mechanisms underlying selective estrogen receptor modulators (SERM) and estrogen-dependent regulation of BRCA1 expression. This was a nested match case-control design involving 592 cases and 1171 controls. Two genes upstream of BRCA1 were discovered (ZNF423 on chromosome 16 and CTSO on chromosome 4) that participate in E2-dependent BRCA1 expression. These biomarkers may assist in individualizing SERM therapy; furthermore, they also provide a potential novel mechanism for individual variation in SERM response. These findings provide strong basis for future clinical research.
Regarding AI therapy, substantial research has focused on the aromatase gene CYP19. Prof. Ingle and colleagues are conducting a GWAS in Patients Experiencing Recurrence of Breast Cancer While Receiving AIs for Early Breast Cancer on NCIC CTG [National Cancer Institute of Canada Clinical Trials Group] trial MA.27 (anastrozole vs exemestane). Over 75% (5221 of 6827) of the enrolled North American patients will be genotyped as part of this study.
Matthew J. Ellis, MD, PhD, Washington University School of Medicine, St. Louis, Missouri, USA, presented a review of whole genome sequencing for luminal-type breast cancer [Ellis MJ et al. Nature 2012].
Whole genome sequencing is an unbiased approach to detect all classes of somatic mutations in a cancer (eg, single nucleotide variants, small insertions and deletions, structural variations). It provides digital data, or information on the frequency with which a somatic variant is observed.
Dr. Ellis and colleagues applied this technique to correlate the variable clinical features of estrogen receptor (ER)-positive breast cancer with somatic alterations. Pretreatment tumor biopsies from patients in two studies of neoadjuvant AI therapy were utilized. Eighteen significantly mutated genes were identified; these included 5 genes previously linked to hematopoietic disorders (RUNX1, CBFB, MYH9, MLL3, and SF3B1). Dr. Ellis concluded that breast cancer, like leukemia, may be viewed as a stem cell disorder that produces indolent or aggressive tumors with various phenotypes depending on differentiation blocks generated by different mutation repertoires.
Mutant MAP3K1 was associated with LUM A status, low-grade histology, and low proliferation rates; mutant TP53 was associated with the opposite pattern. Mutant GATA3 correlated with suppression of proliferation with AI treatment. Mutations in MAP2K4 produced similar permutations as MAP3K1 loss.
Despite enormous complexity, Dr. Ellis and colleagues were able to define frequently mutated genes with significant effects on the clinical phenotype of (ER)-positive breast cancer. Furthermore, bioinformatics analysis identified a number of key genes (functional “hubs”) as consistent features of AI-resistant tumors (eg, MYC, FYN, MAP kinases); targeting these hubs could produce positive outcomes.
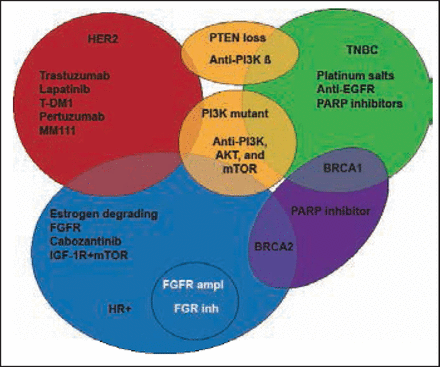
Jose Baselga, MD, PhD, Harvard Medical School and Massachusetts General Hospital, Boston, Massachusetts, USA, discussed new targeted agents in luminal breast cancer. In recent years, the advances in identification of genetic alterations and signaling pathways driving cancer progression, together with the well-defined molecular subtypes of breast cancer have led to the development and evaluation of molecular-targeted therapeutic agents. LUM B is of particular interest due to its increased resistance to hormonal therapy. Inhibitors of the PI3K-AKT-mTOR pathway have recently been explored (Figure 1).
Targeted Therapies for Breast Cancer Subtypes.
Adapted from Higgins MJ, Baselga J. Targeted Therapies for Breast Cancer. J Clin Invest 2011;121(10): 3797–3803.
The Phase 3 BOLERO-2 [NCT00863655] trial, which enrolled 724 patients with ER-positive, advanced breast cancer refractory to nonsteroidal AIs, evaluated exemestane alone at 25 mg QD (n=239) compared with exemestane (25 mg/day) plus everolimus (10 mg/day) [Baselga J et al. N Engl J Med 2012]. The combination therapy resulted in marked improvement in progression-free survival compared with exemestane alone (6.9 months vs 2.8 months; HR, 0.43; 95% CI, 0.23 to 0.54; p<0.001). Additionally, the clinical benefit achieved in the combination arm far exceeded that of the single-agent everolimus in a similar patient population.
Newer, more potent PI3K inhibitors are under clinical development. These include PI3Kα inhibitors, such as BYL719 [Juric D et al. AARC 2012. Abstract CT-01], that have shown activity in PI3Kα mutant tumors patients with HR-positive breast cancer harboring PI3K mutations. Other agents under evaluation include MEK pathway inhibitors and tyrosine kinase inhibitors. Development of these agents will require a focus away from the current large randomized trials in unselected patient populations, in favor of smaller, more targeted patient populations with molecularly defined tumor types. New challenges include the development of resistance due to acquired secondary mutations or induction of adaptive activation of compensatory pathways that prevent cell death. It is likely that combination therapies which act on the secondary mutations and/or the compensatory pathways will significantly improve on the overall effects of targeted agents used alone.
In conclusion of this session, studies are urgently needed to elucidate mechanisms of late relapse in LUM A. Development of new agents to delay or reverse resistance to endocrine therapy— with a truly “targeted” approach—is a priority for LUM B. PGx in endocrine therapy have not provided meaningful deliverables to patients thus far. However, PGx studies have identified new biology that provides clear direction for further translational research that has the potential to substantially benefit patients. PGx is part of the process of scientific discovery.
Additional research utilizing well-characterized cohorts is warranted. Ideal future genomic studies should include prospective informed consent, samples of high quality, and detailed outcomes from clinical trials that address important clinical phenotypes and response to treatment. Low-frequency mutations still represent a substantial number of patients and should be the focus of future clinical trials; these will require comprehensive genome sequencing and large mutation screening programs.
The editors would like to thank the many members of the ESMO Congress 2012 presenting faculty who generously gave their time to ensure the accuracy and quality of the articles in this publication.
- © 2012 MD Conference Express®