Summary
Genetic testing for variants of inosine monophosphate dehydrogenase isoforms will not predict clinical response to mycophenolate mofetil (MMF) in the treatment of lupus nephritis. The Study of Mycophenolate Mofetil in Management of Patients with Lupus Nephritis [ALMS] demonstrated the efficacy of MMF for induction and maintenance of response in lupus nephritis, but response was not uniform, leading to a search for predictors of efficacy.
- Rheumatology Clinical Trials
- Lupus
Genetic testing for variants of inosine monophosphate dehydrogenase (IMPDH) isoforms will not predict clinical response to mycophenolate mofetil (MMF) in the treatment of lupus nephritis.
The Study of Mycophenolate Mofetil in Management of Patients with Lupus Nephritis [ALMS] demonstrated the efficacy of MMF for induction and maintenance of response in lupus nephritis, but response was not uniform, leading to a search for predictors of efficacy. The work was supported by a National Institutes of Health grant (5R01AR55088 to Robert Clancy).
Noa Schwartz, MD, New York University, New York, New York, USA, described MMF as a prodrug of mycophenolic acid (MPA), an inhibitor of IMPDH. IMPDH is the rate-limiting enzyme in the de novo biosynthesis of guanine that drives inosine monophosphate to xanthosine monophosphate with the reduction of nicotinamide adenine dinucleotide (NAD) to NADH. Inhibiting IMPDH prevents the formation of guanine, a crucial component of DNA and RNA through the de novo pathway.
The pathogenesis of systemic lupus erythematosus (SLE) is based on the hyperactivity of both T cells and B cells. Lymphocytes are dependent solely on the de novo pathway for purine production—the same pathway that is inhibited by MMF. It is thought that MMF's inhibition of purine production prevents overstimulated SLE lymphocytes from overproliferating, thus attenuating inflammation as well as disease progression.
MMF has been found to affect other aspects of the inflammatory process. By impairing the production of guanosine triphosphate, MMF attenuates the pathway leading to nitric oxide (NO) production. Therefore, it might be possible to assess the effectiveness of mycophenolic acid at the molecular level by measuring serum NO.
The 2 known isoforms of IMPDH are IMPDH 1, a constitutive enzyme found in most cell types, and IMPDH 2, which is inducible and found mostly in lymphocytes. Both isoforms have many single nucleotide polymorphism (SNP) variations. One specific SNP of each was chosen to study: the rs2278294 from IMPDH1 and rs11706052 from IMPDH2. Both candidate SNPs have been described as being significant in predicting clinical response to MMF.
The IMPDH1 SNP has been found to be inversely associated with the incidence of biopsy-proven acute rejection post-transplantation [Wang J et al. Clin Pharmacol Ther 2008; Gensburger O et al. Pharmacogenet Genomics 2010]. A variation in the IMPDH2 SNP associates with a higher risk of acute rejection in renal transplant recipients [Winnicki W et al. Pharmacogenomics J 2010; Grinyó J et al. Transpl Int 2008]. Its presence is therefore thought to diminish the effect of MMF.
To determine whether the variants of IMPDH associate with clinical response to MMF as well as produce the expected molecular response in subjects enrolled in the ALMS study, DNA and blood samples were obtained from 66 participants for evaluation of genetic variation and serum NO levels. Patients were stratified into 2 groups: Group 1 consisted of responders to MMF at induction and nontreatment failures at maintenance, and Group 2 consisted of nonresponders at induction and treatment failures at maintenance.
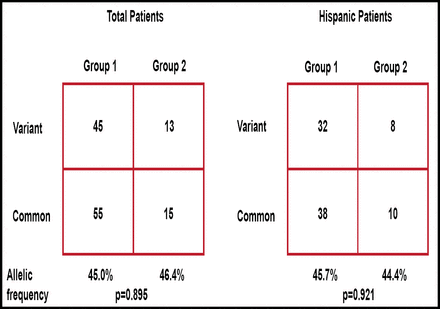
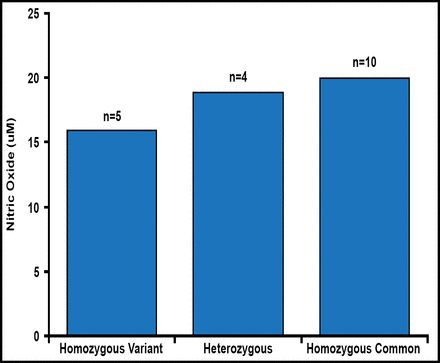
The distribution of the variant alleles for IMPDH1 and IMPDH2 were similar in all subjects regardless of their eventual response to MMF therapy. For the IMPDH1 rs2278294, the distribution of variant alleles was similar in Groups 1 and 2 (allelic frequency, 45.0% vs 46.4%, respectively; Figure 1). Similar results were observed when the analysis was restricted to Hispanics. For the IMPDH2 rs11706052, no association was observed between Groups 1 and 2 (allelic frequency, 7.8% vs 7.1%). There was also no association between genotypes at rs2278294 and NO levels (Figure 2).
Distribution of Variant Alleles for the IMPDH1 Variant rs2278294.
IMPDH1=inosine monophosphate dehydrogenase.
Nitric Oxide Levels Independent of Genetic Variation at IMPDH1.
IMPDH1=inosine monophosphate dehydrogenase; NO=nitric oxide.
Reproduced with permission from N Schwartz, MD.
The data indicate that genetic variations at IMPDH1 and IMPDH2 do not account for the variability in response to MMF nor are they reflected in serum NO levels. As such, genetic testing of these alleles cannot be recommended for predicting the efficacy of MMF in lupus nephritis.
- © 2012 MD Conference Express®