Summary
This article discusses the Cohorts for Heart and Aging Research in Genomic Epidemiology, pharmacogenomics, genomics analyses and Mendelian randomization, as well as the value of genomic analyses carried out in diverse populations in revealing disease variation.
- cardiology genomics
CHARGE CONSORTIUM
Bruce M. Psaty, MD, PhD, University of Washington, Seattle, Washington, USA, discussed the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE). The consortium was formed to spur genome-related research, such as meta-analyses and large longitudinal cohort studies of cardiovascular disease and aging involving many phenotypes.
The genesis of CHARGE was a genome-wide association study (GWAS)—the Cardiovascular Health Study, involving 4056 participants recruited from 1989 to 1990—assessing if genetic mutations common to the participants related to myocardial infarction, heart failure, and stroke. The study highlighted the need for large numbers of patients and controls beyond the scope of any individual study to definitively determine a causal link between genetic mutations and disease. A collaborative approach was necessary.
CHARGE originally consisted of 5 studies encompassing multiple common phenotypes among approximately 38 000 patients [Psaty BM et al. Circ Cardiovasc Genet. 2009]. The organizational structure involves steering, analysis, and genotyping committees, as well as about 40 phenotype-specific working groups. The consortium meets biannually. Analyses within studies involve additive genetics—assessing how a genetic change influences the disease of concern. Prospectively planned meta-analyses are used to examine cohorts from different studies [Skol AD et al. Nat Genet. 2006]. More recently, the consortium has begun merging study databases to allow whole genome sequence-based analysis of large populations [Morrison AC et al. Nat Genet. 2013]. The approach, which calls for trust and transparency among research groups, represents a scientific commons.
DISCOVERING DISEASE VARIATION
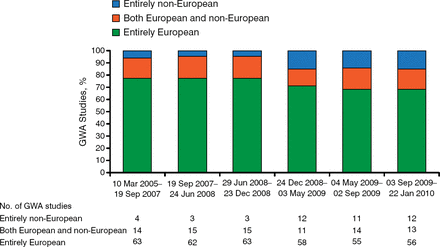
Donna K. Arnett, PhD, University of Alabama at Birmingham, Birmingham, Alabama, USA, talked about the value of genomic analyses carried out in diverse populations in revealing disease variation. Studying different populations rather than 1 ethnic population carries several advantages—for instance, different risk variants occur and can have different effect sizes among populations. Despite this knowledge, the bulk of GWAS data in recent years have come from Caucasian populations (Figure 1) [Rosenberg NA et al. Nat Rev Genet. 2010].
Populations Represented in Genome-Wide Association Studies
GWA, genome-wide association.
Adapted by permission from Macmillan Publishers Ltd: Nat Rev Genet. Rosenberg NA et al. Genome-wide association studies in diverse populations. 2010;11:356–366. Copyright 2010.
The narrow focus has allowed the development of common-variant single-nucleotide polymorphism assays, exploitation of existing data, and use of shared control populations and has permitted meta-analyses—all laudable achievements. However, it is increasingly evident that genomic data from 1 ethnicity cannot be generalized to other ethnicities. This has been exemplified by Dr Arnett's 2 decades of research concerning left ventricular hypertrophy, which is more prevalent and heritable in African Americans with hypertension when compared with hypertensive Caucasians [Arnett DK et al. Am J Hypertens. 2001].
A productive approach has been to select genes that display consistent findings in different species. This has led to the identification of genes with a purported association with obesity, diabetes mellitus, mammary tumor development, and cholesterol regulation. A current focus of the Hypertension Genetic Epidemiology Network, headed by Dr Arnett, is the sequencing of regulatory regions of genes of interest in hypertensive African Americans, with the goal of better understanding the basis of the increased prevalence of left ventricular hypertrophy in this population.
GENOMICS ANALYSES, MENDELIAN RANDOMIZATION, AND THE EVOLUTION OF CARDIOMETABOLIC RESEARCH
Juan Pablo Casas, MD, PhD, University College London, London, United Kingdom, discussed the contributions of genomics analyses and Mendelian randomization—the use of gene variation to assess the influence of modifiable exposure on disease—to advancements in research on cardiometabolic disorders.
The need for innovative approaches has become more pressing as drug development has continued to wane over the past half-century [Scannell JW et al. Nat Rev Drug Disc. 2012]. When development is discontinued late in the development process—as in following a phase 3 randomized controlled trial where the most crucial evidence is generated—the time (a decade or more) and money (tens of millions of dollars) spent are wasted. This has prompted interest in a genomics-oriented approach, which may allow acquisition of randomized evidence in a manner that mimics the trial-driven drug development, without the need for years of research and a level of funding that can be difficult to absorb when development proves unsuccessful.
While a drug intervention trial imposes randomization, a genetic association study relies on natural randomization, in which genetic mutations associated with the target disorder that already exist in a population can be discovered. The relevance of mutations to the risk of development of the particular disorder or disease can be determined. By genomically ruling out certain targets and implicating other targets, traditional drug development can be streamlined. For example, this approach table might have been useful in the VISTA-16 phase 3 trial, which assessed the efficacy of the secretory lipase inhibitor varespladib in the treatment of acute coronary syndrome and which was stopped in 2012 owing to futility. A subsequent Mendelian randomization study determined that genetic variants associated with reduced levels of the target of varespladib were not cardioprotective [Holmes MV et al. J Am Coll Cardiol. 2013]. Had the genomic analysis been done during drug development, the phase 3 trial might never have been done.
Finally, genomics and Mendelian randomization can be valuable in distinguishing effects based on the intended drug-target interaction versus unwanted effects based on drug interactions with other targets and in revealing other applications for an existing drug (eg, using tocilizumab for treatment of coronary heart disease) [IL6R MR Consortium. Lancet. 2012].
PHARMACOGENOMICS
Julie A. Johnson, PharmD, University of Florida, Gainesville, Florida, USA, discussed pharmacogenomics, which studies the role of genetics in drug response. The approach has several goals (Table 1).
Goals for Pharmacogenomics
Because the effect sizes can be much larger in pharmacogenomics studies than in traditional disease genetics studies, with odds ratios of 1.8 to 4.0 being not uncommon, the number of samples needed to detect a disease association can be much smaller, which can speed the timing of clinical implementation. A recent example involved the use of pharmacogenetic and clinical data to estimate warfarin dose [International Warfarin Pharmacogenetics Consortium. N Engl J Med. 2009]. The pharmacogenetics approach has also allowed for comprehensive study of ethnicity-related variation in warfarin dosing [Perera MA et al. Lancet. 2013] and treatment outcomes in manageable numbers of subjects.
Pharmacogenetic data such as these and the demonstrated value of the use of single-nucleotide polymorphisms to define the risk of therapy in different ethnicities [McDonough CW et al. Hypertension. 2013] are reinforcing the view that models and approaches developed for Caucasians are not generalizable to other populations. Pharmacogenetic information may one day guide drug selection in a patient-tailored manner, with resulting better outcomes.
- © 2014 MD Conference Express®