Summary
Colorectal cancer (CRC) was one the first solid malignancies to be classified using molecular profiling, and led to the identification of critical genes and pathways involved in this disease. This article discusses future directions of biomarker research, the molecular biomarker characterization and subtyping of CRC, and the implications of tumor heterogeneity.
- Oncology Genomics
- Gastrointestinal Cancers
- Oncology Genomics
- Oncology
- Gastrointestinal Cancers
Colorectal cancer (CRC) was one the first solid malignancies to be classified using molecular profiling, and led to the identification of critical genes and pathways involved in this disease. An Education Session at the ASCO 2014 Annual Meeting, “The Evolution of Our Molecular Understanding of Colorectal Cancer: What Are We Doing Now, What the Future Holds, and How Tumor Profiling is Just the Beginning,” concerned the current status of CRC biomarkers and the challenges facing the development of effective treatment because of CRC heterogeneity.
CURRENT COLORECTAL CANCER BIOMARKERS
Sabine Tejpar, MD, PhD, University of Leuven, Belgium, presented “What Is the Current Status of Biomarkers in Use for the Treatment of Patients with Colorectal Cancer?” and speculated on the future directions of biomarker research. The current CRC biomarkers include RAS mutations and microsatellite instability (MSI) or deficient mismatch repair (dMMR). MSI is a good prognostic factor in stage II CRC, and its presence may allow patients to avoid adjuvant chemotherapy. Dr. Tejpar reviewed the structure of KRAS and NRAS. Mutations in the second exon of KRAS are the most common; if KRAS exon 2 is wild-type, then testing for other KRAS or NRAS mutations is justified and necessary for decision making regarding the use of anti-EGFR therapy.
Prof. Tejpar emphasized that CRC is biologically heterogeneous and indicated that a subgroup analysis of clinical trial results can reveal patient subgroups for which therapies, including targeted agents, have different efficacies or may be harmful. Numerous studies have defined different CRC subgroups or molecular subtypes. For example, the Cancer Genome Atlas Network [Nature 2012] performed a genome-scale analysis of 224 CRC and normal tissue pairs, identifying new mutations and showing that more than 90% of both hypermutated and non-hypermutated tumors have mutations in 1 or more members of the WNT signaling pathway, and that hypermethylation was more common in tumors from the ascending (right) colon, suggesting that CRC is not one but many diseases affecting the same organ. In another study, DNA methylation patterns were used to define 4 methylation subgroups of CRC [Hinoue T et al. Genome Res 2013]. The multinational Colorectal Cancer Subtyping Consortium (CRCSC) aims to reconcile the disparate findings from research groups searching for molecular subtypes of CRC. Novel target discovery must address differences within and between tumors, the contribution of the intestinal flora, and the roles of mutations, methylation patterns, among others in this disease.
MATCHING TARGETED THERAPIES
Josep Tabernero, MD, PhD, Vall d'Hebron University Hospital, Barcelona, Spain, session chair, presented “Molecular Biomarker Characterization and Subtyping of Colorectal Cancer: Next-Generation of Matched Targeted Therapies” and reiterated much of what Dr. Tejpar presented. He agreed that although the molecular understanding of CRC is quite advanced compared with that of other tumor types, the correlation among published studies of gene expression profiling in CRC is limited.
Prof. Tabernero recommends using the mutational profile of KRAS, NRAS, BRAF, PIK3CA and MSI status for stratifying patients in clinical trials of matched targeted therapies. One benefit is the ability to avoid the detrimental effect of anti-EGFR monoclonal antibodies in patients with specific tumor mutations. So-called liquid biopsies or the identification of the DNA from circulating tumor cells in the blood (ctDNA) should make it easier to identify primary and acquired mechanisms of resistance to targeted therapies. Future directions include developing CRC molecular subtypes based on gene expression profiling that could be used to develop personalized therapy.
IMPLICATIONS OF TUMOR HETEROGENEITY
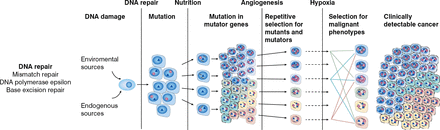
Charles Swanton, MD, PhD, University College London Cancer Institute, London, United Kingdom, gave a talk titled “What is Tumor Heterogeneity and How Will This Play a Role in the Development of Agents to Treat Colorectal Cancer?” Tumor heterogeneity in CRC affects the ability to validate biomarkers and increases the cost of drug development. Known drivers of CRC include defects in DNA repair mechanisms, epigenetic changes, and chromosomal instability (Figure 1).
Drivers of Tumor Heterogeneity in Colorectal Cancer
CIMP=CpG (cytosine-guanine dinucleotide) island methylator phenotype; CRC=colorectal cancer.
Reproduced from Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer 201111(6):450–457. With permission from the Nature Publishing Group.
Early events in CRC tumor evolution, which are present in every cancer cell, include KRAS and BRAF mutations. Tumor heterogeneity also results in acquired drug resistance and tumor metastasis. Drug resistance is almost inevitable, and multiple resistant genetic events evolve over time within individual patients. Drug development strategies therefore need to be based on an understanding of the mechanisms driving tumor diversity, possibly by targeting the diversity or by limiting its development.
Drivers of chromosomal instability (CIN) in CRC are not completely understood. Dr. Swanton's group looked at patterns of chromosomal segregation errors that lead to CIN in models of CRC. In aneuploid CRC tumors and CIN cell lines, a region of chromosome 18q is frequently lost early in the adenoma–carcinoma transition and is more frequent in carcinomas. Knocking out each of the 3 genes identified in this region increased chromosomal instability, induced structural damage, and replication stress. Although the addition of nucleosides at least partially rescued this replication-induced DNA damage in cells, this was not suggested as a treatment strategy but as a means of understanding the process.
Dr. Swanton went on to discuss the role of genome doubling in CRC genome evolution, a phenomenon that occurs commonly in nature; for example, in plants adapting to stress. Using a CRC cell line, few single cellcloned tetraploid cells survive, and most single cellcloned diploid cells survive, suggesting a major hurdle in tolerating the genome doubling event [Dewhurst SM et al. Cancer Discovery 2014]. However, after propagating these clones over 2 years, the diploid cells tended to die or arrest during segregation, whereas the tetraploid cells tolerated segregation errors and further heterogeneity well and continued to propagate. At the 18-month time point, for example, cultured diploid cells remained genetically stable, whereas genome instability accelerated in the tetraploid cells, which drifted toward triploidy and increased instability. These findings suggest that the tetraploid event buffers CRC cells so that they can drift towards an adaptive state with enhanced fitness and aggressiveness. Indeed, in tumors, the onset of genome doubling was associated with a higher risk of relapse in early-stage CRC [Dewhurst SM et al. Cancer Discovery 2014].
In summary, the identification of certain CRC driver mutations has allowed patients to receive matched targeted therapies or to avoid inappropriate treatment. However, CRC is a complex, molecularly heterogeneous disease; therefore, a classification system that defines a personalized approach to therapy for individual patients will be instrumental for the more accurate tailoring of treatments. In addition, future therapeutic approaches will need to consider genomic instability and the drivers of tumor heterogeneity.
- © 2014 MD Conference Express®