Summary
The rapid emergence of antibiotic resistance is a major public health concern [Zhang L et al. Appl Environ Microbiol 2011]. This article discusses five questions of resistant bacteria in the gut: Their possible presence without antibiotic exposure; Whether there is selection of resistant gut bacteria during antimicrobial exposure; Whether there is selection of resistance during systemic treatment for other infections; Whether it is possible to avoid or minimize selection; How optimization of treatment relates to selection of resistance in the gut.
- Bacterial Infections
- Drug Resistance
Resistance in the Gut
The rapid emergence of antibiotic resistance is a major public health concern [Zhang L et al. Appl Environ Microbiol 2011]. Johan W. Mouton, MD, Nijmegen Institute for Infection, Inflammation & Immunity, Nijmegen, The Netherlands, discussed five questions of resistant bacteria in the gut:
-
Their possible presence without antibiotic exposure
-
Whether there is selection of resistant gut bacteria during antimicrobial exposure
-
Whether there is selection of resistance during systemic treatment for other infections
-
Whether it is possible to avoid or minimize selection
-
How optimization of treatment relates to selection of resistance in the gut
Data suggest that early development of antibiotic resistance in human gut microbiota is independent of an infant's exposure to antibiotics but is likely to be affected by exposure to maternal and environmental microbes during and after delivery. The population of food-borne antibiotic-resistant bacteria is also significantly amplified within the host, even in the absence of antibiotic-selective pressure [Zhang L et al. Appl Environ Microbiol 2011].
Prof. Mouton cited a study in which 2 of 20 children with no known antibiotic exposure, living in a very remote Senegalese village, were fecal carriers of a multiresistant Escherichia coli clone that produced CTX-M-15 [Ruppe E et al. Antimicrob Agents Chemother 2009], strongly suggesting that the pC15–1a multidrug-resistant region can persist in the intestinal flora in the absence of significant selective pressure, at least that we know of.
Based on a report by de Smet et al. [Lancet Infect Dis 2011], Prof. Mouton justified the widespread use of selective digestive tract decontamination in intensive care units with low levels of antibiotic resistance. Prof. Mouton presented an extensive analysis of an experimental study that looked at the effects and duration of antimicrobial treatment for pneumonia in selecting resistant microorganisms in the gut [Goessens WH et al. JAC 2007]. This showed that the more frequent the dosing regimen, the higher the propensity for selecting resistant bacteria. Emergence of resistance is dependent on dose (inverse U shape), duration of therapy, and dosing regimen. For the first three questions that were posed, Prof. Mouton answered “yes;” “perhaps” to the fourth; and “not good” to the fifth.
Resistance in a Dynamic Model
Didier Guillemot, MD, Institut Pasteur/Univ. Versailles Saint Quentin/Inserm, Paris, France, discussed the impact of antibiotic dose on resistance selection in the community. His findings were based on a dynamic model of Streptococcus pneumoniae. His presentation covered β-lactam doses and pneumococci susceptibility, accounting for β-lactam doses.
From a public health point of view, antibiotics do more to increase the clearance of susceptible bacteria than the acquisition of a new mechanism or resistant strain. Prof. Guillemot noted that much is known about the relation between S. pneumoniae, antibiotics, and resistance, but not at the population level.
The mathematical model that he discussed assessed the influence of modifying doses of β-lactam at the population level to estimate the impact on resistance levels and prevalence in colonized individuals. Questions that were considered using this model included the effects of prescription frequency, prescribed dose, and whether defined daily dose (DDD) is a good indicator to predict the evolution of β-lactam resistance to S. pneumoniae. Simulations over a 50-year period of fixed- and variable-dose exposure showed a bimodal distribution and that the prevalence of resistance increases with the frequency of exposure. Both findings were consistent with prior epidemiological studies.
Dosing outcomes indicated that higher doses may reduce the prevalence of resistance and increase the minimum inhibitory concentration (MIC) of resistant strains. The model also showed that DDD is not an accurate indicator for predicting pneumococcal resistance to β-lacams. “Don't use DDD to anticipate the future of S. pneumoniae dissemination or to analyze the relationship between antibiotic use and S. pneumoniae resistance,” he said.
Which Matters More — Antibiotic Dose or the Bacterium?
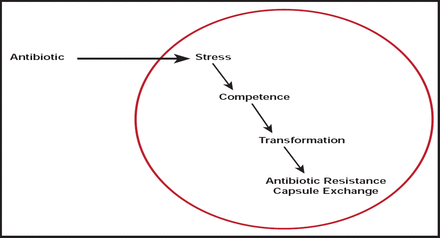
Patrice Courvalin, MD, Institut Pasteur, Paris, France, discussed the relative importance of antibiotic dose or changes in the bacterial genome in causing antibiotic resistance. Bacteria respond to many changes in their environment by sensing small molecules; yet, competence for genetic transformation is transient. In several bacterial species, it depends on achieving a specialized cellular state [Harvarstein LS et al. Proc Natl Acad Sci 1995].
Harvarstein et al. [Proc Natl Acad Sci 1995] found that competence-stimulating peptide induced competence in pneumococcal cultures in a dose-response fashion to the synthetic peptide, with the highest yield (about 5% of cells transformed) observed at doses of 30 to 1000 ng/mL and a monotonic dose response in the intervening region.
Resistance in Acinetobacter spp., particularly Acinetobacter baumannii, provides another example. A. baumannii possesses two intrinsic β-lactamase genes, in addition to weak permeability and efflux systems. Together, they confer a natural reduced susceptibility to antibiotics. Numerous acquired mechanisms of resistance and genetic elements, such as resistance islands, have also been identified [Poirel L et al. IUBMB Life 2011].
Based on these and other findings, Prof. Courvalin concluded that antibiotics promote evolution of resistance (Figure 1).
Antibiotics Promote Evolution of Resistance.
Reproduced with permission from P. Courvalin, MD.
PK-PD and Resistance
William A. Craig, MD, University of Wisconsin School of Medicine and Public Health, Madison, Wisconsin, USA, discussed the use of pharmacodynamics/pharmacokinetics (PD/PK) to establish the target that is required to prevent an increase in resistant populations; to identify which PK/PD indices (Cmax, AUC/MIC, T>MIC) or other characteristics best prevent the emergence of resistance; and to determine the magnitude of the PK/PD indices or other characteristics that is required to prevent the development of resistance.
With regard to the mutant prevention concentration (MPC) that stops mutant selection at 1010 organisms, he reported that MPC is usually 2- to 16-fold higher than MIC, with selection of resistance higher if drug concentrations persist in the zone between the two concentrations [Blondeau JM et al. Antimicrob Agents Chemother 2001]. He also pointed out the inverted U-shaped distribution of resistance emergence versus dose intensity [Tam VH et al. Antimicrob Agents Chemother 2007].
Dr. Craig discussed aminoglycide dosing to minimize resistance for Enterobacteriaceae and Staphyloccocus aureus (Cmax/MIC >6 [once-daily dosing]) and the need for a Cmax/MIC of 30 with twice-daily dosing of gentamicin to prevent emergence of resistance with P. aeruginosa [Tam VH et al. Antimicrob Agents Chemother 2008].
In addition, he covered PK/PD indices that are associated with in vitro enhancement or suppression of fluoroquinolone resistance, including AUC24/MIC, AUC24/MPC, and Cmax/MIC, and how doxycycline, combined with moxifloxacin, can reduce emergence of resistant S. aureus [Allen GP, Deshpande LM. Int J Antimicrob Agents 2010]. Dr. Craig concluded that there is a need for more in vivo studies on optimal dosing and combination therapy to effectively prevent resistance.
- © 2011 MD Conference Express