Summary
The results of genome-wide association studies and functional genomics studies reviewed in this article have elucidated new biology in breast cancer that provides a foundation for further laboratory and clinical research to identify genetic variability and its effects on breast cancer prevention and treatment.
- treatment strategy
- prevention
- genetic variability
- functional genomics study
- genome-wide association study
Pharmacogenomics (PGx) can be harnessed to develop and guide treatment that is tailored to the characteristics of an individual patient with breast cancer (BC) and to identify strategies to prevent BC. James N. Ingle, MD, Mayo Clinic, Rochester, Minnesota, USA, reviewed the variability in the hormonal milieu in response to endocrine therapy (ET); genome-wide association studies (GWASs), which give insights into the statistical significance of genetic variability; and functional genomics studies, which give insights into mechanisms, in the William L. McGuire Memorial Lecture.
GWASs are an unbiased way to look across the genome for genetic variability related to a specific phenotype, and they can lead to the discovery of new biology relating to genes and mechanisms in ET of BC, stated Dr Ingle. The estrogen receptor (ER) is a transcription factor that binds to an estrogen response element (ERE), which is a sequence of DNA that regulates transcription and has a palindromic motif. Single-nucleotide polymorphisms (SNPs) can create or disrupt EREs. Surprisingly, SNPs that are hundreds of base pairs away can have an effect on EREs. Equally surprisingly, tamoxifen (TAM) can reverse SNP-dependent effects on ERα binding to EREs.
The variability in patient response to ET includes differences in disease outcomes, musculoskeletal (MS) events with aromatase inhibitors (AIs), and deep vein thrombosis, lipid effects, and hot flashes with TAM. Genetic variability plays a role in this clinical variability in response, in part through alterations in the pharmacodynamics and pharmacokinetics of the drugs, calling for consideration of the need to adjust drug doses for specific patients.
In a study of the PGx of AIs in postmenopausal stage I to III hormone receptor–positive BC treated with anastrozole 1 mg daily, there was a 90% reduction in the estradiol levels in only 79% of the 643 patients [Ingle JN et al. Steroids. 2014]. This observation led to a GWAS of baseline estradiol, which implicated SNPs near the TSPYL5 genome in the levels of estradiol, with a median twice the higher level [Liu M et al. Mol Endocrinol. 2013]. Estradiol induced TSPYL5 expression, and TSPYL5 induced CYP191A (aromatase) expression, thereby creating a positive feedback loop. Furthermore, TSPYL5 is bound to the CYP191A I.4 promoter gene. The large variability in anastrozole concentrations observed in vitro and in vivo [Ingle JN et al. Cancer Res. 2010; Kamdem LK et al. Br J Clin Pharmacol. 2010] led to a GWAS that found 2 signal genes that were significantly correlated with genes encoding transporters, CYP450 enzymes, and UGT enzymes in hepatic tissue [Dudenkov TT et al. ISSX 2014 (poster 3710)]. Based on the results of these studies, ongoing research includes a GWAS with the phenotype of change in hormones and development of a panel of SNPs that will be studied in 2 large clinical trials of adjuvant AI therapy.
A PGx study was also conducted using the DNA from women in North America with early BC treated with adjuvant AI therapy in the NCIC CTG MA.27 study, which found no difference between exemestane and anastrozole for BC outcomes at 5 years [Goss PE et al. J Clin Oncol. 2013]. MS adverse events (AEs) are the primary reason for discontinuing AIs, and a GWAS identified the TCL1A gene and the interleukin-17 (IL-17) cytokine to be associated with MS AEs in women treated with AI therapy [Ingle JN et al. J Clin Oncol. 2010]. Further work found an SNP-dependent difference in TCL1A expression and altered the expression of cytokines, NF-κB, and chemokines, which may help to explain the MS AEs in some women taking AI [Liu M et al. Breast Cancer Res. 2012]. Ongoing work is searching for other inflammatory mediators that may be druggable, deep sequencing across the T-cell leukemia gene cluster, and an analysis of a GWAS with breast events (ie, breast recurrence) as the phenotype.
Prevention of Breast Cancer
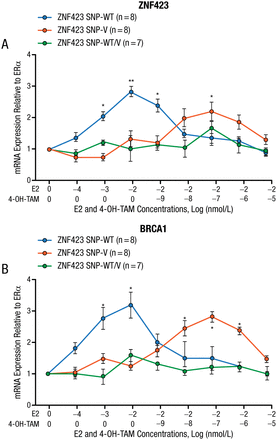
The best hope for making major advances against BC may be preventing it because of the chaotic mutations in tumors, stated Dr Ingle. Six trials evaluated the selective estrogen receptor modulators (SERMs) TAM and raloxifene as preventive agents in women at high risk of developing BC, and 2 of these trials (National Surgical Adjuvant Breast and Bowel Project [NSABP] P-1 and P-2) included almost 33 000 women, which represents 59% of the world’s experience. In women who received a SERM on NSABP P-1 and P-2, DNA samples from 592 cases (women who developed BC) and from 1171 controls (women who did not develop BC) were used in a GWAS that found variant SNPs in a gene (ZNF423) that were protective and variant SNPs near a gene (CTSO) that were deleterious [Ingle JN et al. Cancer Discov. 2013]. Women who were homozygous for both deleterious alleles had an odds ratio of 5.71 for developing BC compared with those women who were homozygous for both protective alleles. This suggests the possibility of identifying a population of patients for a prospective prevention study, stated Dr Ingle. Furthermore, in laboratory studies using lymphoblastoid cell lines of known genotype, they showed that cells with the wild-type (WT, deleterious) ZNF423 SNP had increasing ZNF423 and BRCA1 expression with increasing doses of estradiol, but that when there was ERα blockade with a SERM, the expression of ZNF423 and BRCA1 was “reversed” with decreased expression with the WT and increased expression with the variant SNP (Figure 1).
mRNA Expressions
Error bars represent SEM.
4-OH-TAM, 4-hydrotamoxifen; E2, estradiol; ER, estrogen receptor; OnME2, zero nano molar estradiol; SEM, standard error of the mean; SNP, single-nucleotide polymorphisms; V, variant; WT, wild type.
*P < .05 vs OnME2.
**P < .01 vs OnME2.
Adapted from Cancer Discovery, Copyright 2013;3:812–825, Ingle JN et al, Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. With permission from AACR.
The variant ZNF423 SNP that increased BRCA1 expression after ERα blockade was 200 base pairs away from the ERE that affected ERα binding, and a series of laboratory studies was performed to understand this mechanism. The current working model is that coiling of the chromosome allows the SNP to be in closer proximity to the ERE and affect binding. Ingle and colleagues continue to work on a protein that will mediate the SNP-dependent difference in ERα binding to the ERE in the presence of 4OH-TAM.
Dr Ingle stated that these findings provide focus for further research and that functional studies are ongoing. He called for a clinical trial to test the hypothesis that the genetic variants identified are related to efficacy of SERM therapy in women at high risk of developing BC, and at the same time for collecting comprehensive biospecimens that can be used for multiple “pharmaco-omic” studies, including metabolomics, lipidomics, proteomics, transcriptomics, epigenomics, and the microbiome.
Conclusions
There is substantial interindividual variability in sex hormones in women who are postmenopausal and in the response to anastrozole and SERMs. SNPs can create or disrupt an ERE and can influence binding, even from a distance of hundreds of base pairs from the ERE. SERMs can reverse ERα binding to an ERE in an SNP-dependent fashion. GWASs are an important discovery tool in a process that examines SNP function and relationship to genes, drug effects, and clinical phenotypes. PGx has identified new biology that provides direction for further laboratory and clinical research, and it has substantial potential for meaningful benefit to patients with further work.
- © 2014 SAGE Publications