Summary
Genome-wide association studies (GWASs) of patients with hypertension identified mutations in the uromodulin gene that appear to be associated with blood pressure (BP). This article discusses genomics in relation to BP.
- Hypertensive Disease
- Cardiology Genomics
- Cardiology & Cardiovascular Medicine
- Hypertensive Disease
- Cardiology Genomics
Genome-wide association studies (GWASs) of patients with hypertension identified mutations in the uromodulin (umod) gene that appear to be associated with blood pressure (BP). Anna F. Dominiczak, MD, University of Glasgow, Glasgow, United Kingdom, delivered the Presidential Lecture that discussed genomics in relation to BP.
The underlying cause of essential hypertension is complex and multifactorial, with environmental factors playing a role, such as race, sex, physical activity, caffeine, drugs, diet, and sodium, as well as genetic factors, such as insulin resistance, oxidative stress, sympathetic nervous system, endothelial function, renin-angiotensin-aldosterone system, renal electrolyte transport, and monogenic syndromes [Padmanabhan S et al. J Hypertens 2008]. There are several methods to examine genetic traits that may be involved in hypertension, including candidate gene association studies, genome-wide linkage studies, and GWASs [Simino J et al. Curr Opin Nephrol Hypertens 2012].
In candidate gene association studies, a genotype variant that is expected to affect a trait is tested for an association [Simino J et al. Curr Opin Nephrol Hypertens 2012]. Variants that have a significant association are then validated in functional studies. In genome-wide linkage studies, patients who are related are recruited to the study, and their genomes are analyzed with genotype markers scattered randomly throughout the genome; then, the chromosome region and trait are tested via cosegregation, followed by fine mapping of linkage peaks to localize the specific gene of interest.
In GWASs, an association analysis is performed in cases and controls of a topic of interest, such as hypertension, in which the entire genome is analyzed for single-nucleotide polymorphism (SNP) frequencies [Patel RS et al. Heart 2011]. Top hits enter a validation phase in which an association analysis is repeated in cases and controls. The remaining hits must be replicated in another association analysis or other genotyping technique. These replicated hits should be further verified through functional studies and risk prediction techniques.
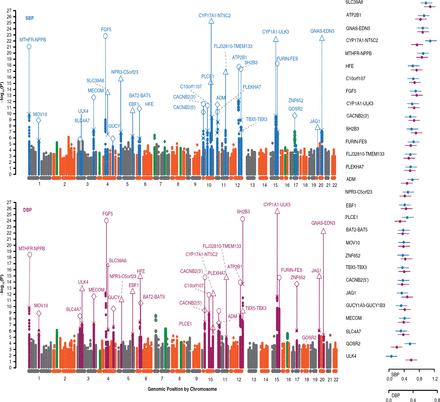
The International Consortium for Blood Pressure-GWAS includes 29 cohorts consisting of up to 70,000 patients, representing the largest GWAS performed in hypertension [Ehret GB et al. Nature 2011]. The validation phase is occurring in European, South Asian, African, and East Asian ancestries. To date, this GWAS has identified multiple SNPs that potentially play a role in systolic or diastolic BPs (SBP; DBP). The Manhattan plots of SNPs and their effect on BP illustrate the change in BP for a given SNP (Figure 1). Prof. Dominiczak pointed out that some critics stated that the change in BP illustrated by these maps was not large enough to translate into a clinical effect. From these identified SNPs, 29 SNPs in aggregate were used to calculate a weighted risk score that was significantly associated with left ventricular wall thickness (p=0.000006), stroke (p=0.000033), and coronary artery disease (p=10 × 10−29) but not chronic kidney disease or kidney function.
Manhattan Plots of the Effect of Single-Nucleotide Polymorphism on Blood Pressure
DBP=diastolic blood pressure; SBP=systolic blood pressure.
Reproduced from Ehret GB et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. With permission from the Nature Publishing Group.
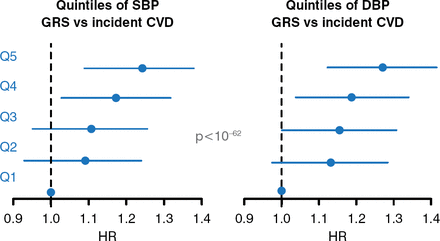
In a Finnish cohort of >32,000 patients, the BP risk score was able to predict the risk of cardiovascular events. Patients in the highest quartile of SBP and DBP had the greatest HR of incident cardiovascular disease, whereas patients in the lowest quartile had a HR of 1.0 (p<10 × 10−62; Figure 2) [Havulinna AS et al. Hypertension 2013].
GWAS-Derived Blood Pressure Risk Score Predicts Incident Cardiovascular Disease
CVD=cardiovascular disease; DBP=diastolic blood pressure; GRS=genetic risk score; GWAS=genome-wide association study; SBP=systolic blood pressure.
Reproduced from Havulinna AS et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32 669 individuals. Hypertension. 2013;61(5):987–994. With permission from Lippincott Williams and Wilkins.
In an attempt to replicate the findings of the Finnish cohort, a GWAS of extremes was conducted—namely, between hypercontrols, who were patients with low BP (≤120/80 mm Hg) with no cardiovascular events over 10 years, and cases, who had 2 consecutive BP readings of 160/100 mm Hg and were diagnosed with hypertension at age <60 years. From these data, the GWAS identified SNP rs13333226 on chromosome 16, which mapped to the umod gene [Padmanabhan S et al. PLoS Genetics 2010]. The identification of this SNP and its association to hypertension was replicated in multiple analysis with a collective cohort size of >39,000 patients, with an overall odds ratio of 0.85 (95% CI, 0.81 to 0.89; p=1.5 × 10−13).
Uromodulin (UMOD) is a large protein located among important transporter proteins in the thick ascending limb of the loop of Henle. The variant G allele of SNP rs13333226 was associated with a lower risk of developing hypertension and lower urinary UMOD excretion. In addition, a direct association between urinary UMOD and sodium excretion was supported by the BRIGHT study in patients with hypertension and in the general population in the HERCULES study. In animal studies, SBP increased in knockout mice with no umod but not mice with wild-type umod that were treated with 2% of sodium chloride by 4 weeks [Graham LA et al. Hypertension 2014].
UMOD protein expression may be altered by some SNPs, or its trafficking may be dysfunctional as a result of SNPs, causing UMOD to be expressed only in the thick ascending limb of Henle [Padmanabhan S et al. 2014]. A decrease in UMOD results in natriuresis occurring at lower elevations in BP.
Prof. Dominiczak highlighted that GWASs can identify new genes and regions of genes involved in hypertension. In terms of the future, >50 candidate genes have been associated with hypertension via candidate gene studies, whereas several replications of various quantitative trait loci identified by genome-wide linkage scans have failed. Despite the promise of GWASs, 29 replicated variants explain only up to 2.5% of BP variance.
Next-generation sequencing (NGS) holds promise to integrate data-intensive biology with medicine [Phimister et al. N Engl J Med 2012]. For example, in clinical trials, an NGS panel could enable investigators to stratify patients into different subtrials according to genetic differences or tumor genetic differences.
Prof. Dominiczak concluded that if genetics, regulatory networks, protein interactions, and metabolic pathways can be brought together, there would be no “secrets” underlying hypertension mechanism in individual patients. This knowledge would have the potential to allow a thorough understanding of the disease phenotype and identify ways in which we could reduce cardiovascular events and stroke.
The editors would like to thank the many members of the 2014 European Society of Hypertension, the Hellenic Society of Hypertension, and the International Society of Hypertension presenting faculty who generously gave their time to ensure the accuracy and quality of the articles in this publication.
- © 2014 MD Conference Express®