Summary
Each year, the American Heart Association presents the Lewis A. Conner Memorial Lecture to honor the memory of one of the founders of the AHA. This year, Joseph Loscalzo, MD, PhD, FAHA, Harvard Medical School, Boston, Massachusetts, USA, delivered a presentation on the potential for a more personal, systems- and network-based approach to drug development.
- Cardiology Genomics
- Prevention & Screening
Each year, the American Heart Association (AHA) presents the Lewis A. Conner Memorial Lecture to honor the memory of one of the founders of the AHA. This year, Joseph Loscalzo, MD, PhD, FAHA, Harvard Medical School, Boston, Massachusetts, USA, delivered a presentation on the potential for a more personal, systems- and network-based approach to drug development.
Standing at the Crossroads
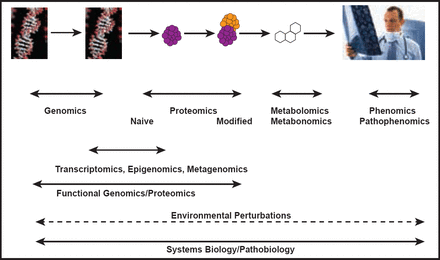
Today, we stand at a crossroads between a one-size-fits-all drug development paradigm and a systems-based approach that incorporates the developing fields of genomics, proteomics, metabolomics, phenomics, pathophenomics, transcriptomics, epigenomics, and metagenomics (Figure 1).
Systems Biology and Pathobiology.
Reproduced with permission from J. Loscalzo, MD, PhD, FAHA.
Integration of systems biology and network analysis is setting the stage for a novel drug development paradigm that is based on the identification of mechanisms that underlie physiological circuits that are corrupted in disease states [Loscalzo J, Barabasi AL. Wiley Interdiscip Rev Syst Biol Med 2011; Waldman SA, Terzic A Clin Transl Sci 2011]. Insights into these processes offer unparalleled opportunities for a network-based approach to human illness [Barabasi AL et al. Nat Rev Genet 2011] and personalized medicine.
The goal of personalized medicine is to match precisely the right drug at the right dose to the specific biology of the disease in an individual patient and to maximize therapeutic efficiency while minimizing adverse and off-target effects [Waldman SA, Terzic A. Clin Transl Sci 2011]. The principle itself is straightforward, but its implementation remains challenging [Hamburg MA, Collins FS. N Engl J Med 2010].
Moving Beyond the Silver Bullet
The science of drug discovery has accelerated exponentially; yet, delivery of its benefits to patients and populations has lagged [Waldman SA, Terzic A. Clin Transl Sci 2008; Waldman SA, Terzic A. Clin Pharmacol Ther 2008]. One reason is that pharmaceutical research and development (R&D) productivity has been in decline since the mid-1990s. This may be due, in part, to an intrinsically flawed reductionist approach to drug development (ie, the need to identify a single drug target or a “silver bullet”).
Since the advent of genomics in the 1990s, potential drugs—typically proteins that appear to have a key role in disease pathogenesis—have been the main focus of discovery efforts. Great advances in the development of new tools to identify targets and compounds that interact with them have taken place. Yet, declines in R&D productivity point to potential limitations in the target-centric approach to drug discovery [Swinney DC, Anthony J. Nat Rev Drug Discov 2011].
Before the introduction of these approaches, drug discovery was driven primarily by phenotype, often with limited knowledge of molecular mechanisms of disease. Nonetheless, the pharmaceutical industry was successful in the discovery and development of innovative medicines [Swinney DC, Anthony J. Nat Rev Drug Discov 2011].
Some suggest that the more limited use of phenotypic screening in recent years has contributed to the current lack of success in drug R&D [Williams M. Biochem Pharmacol 2005; Flordellis CS et al. Curr Top Med Chem 2006]. Others blame an increasing focus of research activities on the development of selective drugs in complex scientific areas that are characterized by a low probability of success.
Between 1998 and 2008, the number of new medical entities (NMEs) that was approved per year declined (although it has been roughly constant since 2005). Conversely, attrition rates, development times, and R&D expenditures have all increased. The growth in attrition rates—the proportion of failures out of the total number of projects that enter any given stage of R&D—has been dramatic [Pammolli F et al. Nat Rev Drug Discov 2011].
The Imperative for Change
From 2000 to 2008, there were 35 NMEs per year out of 1740 new projects. The probability of success was 2.01% per year. Development time was 13.9 years (up from 9.7 years during the 1990s), and United States clinical trials took 7 years on average to complete [Pammolli F et al. Nat Rev Drug Discov 2011]. These statistics represent the status quo—a drug development process that has been in place for the past 50 years [Fishman MC, Porter JA Nature 2005; Butcher EC et al. Nat Biotechnol 2004].
It is widely recognized that major improvements are required in the methods that are now being used to develop new drugs. The time from initial target identification to market can be 10 to 14 years and incur a cost in the hundreds of millions of dollars. Even after substantial investment, only 30% to 40% of the candidate compounds that enter clinical trials are successful [Nordsletten DA et al. IEEE Trans Biomed Eng 2011].
Systems Pathobiology and Network Medicine - The Shape of What is to Come
A contemporary approach to human disease requires that it be viewed from a systems perspective rather than as a simple correlation between clinical syndromes and pathological analysis [Loscalzo et al. Mol Syst Biol 2007; Loscalzo J, Barabasi AL. Wiley Interdiscip Rev Syst Biol Med 2011]. Network modeling of whole-transcriptome expression data, for example, enables characterization of complex gene-gene interactions that underlie cellular functions [Chu J et al. BMC Syst Biol 2011].
Drug-target interactions have been studied in the context of interactome networks or interactions between proteins [Rask-Andersen M et al. Nat Rev Drug Discov 2011; Yidirim MA et al. Nature Biotech 2007]. The field of interactomics represents a new way to understand the success of drug targets in the context of their functional milieu [Rask-Andersen M et al. Nat Rev Drug Discov 2011].
Network-based approaches represent a relatively recent trend in drug discovery. However, the fact that drug development is clearly affected by intricate network effects suggests that network pharmacology will become an essential component of drug development strategies [Schadt EE et al. Nat Rev Drug Discov 2009; Hopkins AL. Nature 2009; Barabasi AL et al. Nat Rev Genet 2011].
According to Dr. Loscalzo, quantitative holistic systems biology that is applied to human disease offers a unique approach for diagnosing established illness, defining disease predilection, and developing individualized (personalized) treatment strategies. These approaches will be able to take full advantage of modern molecular pathobiology and the comprehensive datasets that are rapidly becoming available for populations and individuals, and in so doing, they offer the promise of redefining our approach to disease, its treatment, and the practice of medicine [Loscalzo et al. Mol Syst Biol 2007; Loscalzo J, Barabasi AL. Wiley Interdiscip Rev Syst Biol Med 2011].
- © 2011 MD Conference Express®